- 翰林提供學術活動、國際課程、科研項目一站式留學背景提升服務!
- 400 888 0080
IB DP Biology: SL復習筆記5.2.5 Cladistics
Clades
- The term?clade?can be defined as
- A group of organisms that have all descended from a common ancestor
- Cladistics?is the branch of science in which scientists put organisms into clades
- It involves classification that is based on?homologous characteristics?rather than?analogous characteristics
- Clades are formed on the basis of?evolutionary relationships?i.e. who is descended from which ancestor
- Note that while?taxonomy?is about classifying and then?naming organisms, cladistics is about?identifying evolutionary relationships?between organisms
- A?taxon?is a group of organisms that have been given a group name by taxonomists?on the basis on their shared features
- A?clade?is a group of organisms classified together?on the basis of their shared descent?from a common ancestor
- If taxonomy is carried out correctly then?all of the members of a taxon should form a clade, but due to historical errors and the difficulties in distinguishing between?true homologous characteristics?and those that have come about by?convergent evolution, this is not always the case
- Clades can include both?living?and?extinct?species
- Some of the descendants of a common ancestor may have gone extinct
- The common ancestor species itself may have gone extinct
- Clades can be?large?or?small?depending on the common ancestor being studied
Identifying Members of a Clade
- In the past, scientists encountered many?difficulties?when trying to determine the?evolutionary relationships?between species
- Using the physical features of species has?limitations?and can often lead to organisms being put into?groups that are not true clades
- This would mean that all of the organisms in a group are?not descended from a common ancestor
- Some descendants might be missing
- Some organisms might have been included that descend from a different ancestor
- Advances in?sequencing technology?have allowed scientists to further investigate the?evolutionary relationships between species
- Sequence data?that can be used to investigate evolutionary relationships can come from
- DNA
- mRNA
- Amino acids in polypeptides
- Sequencing technology can determine the?order?of?DNA bases, mRNA bases?and?amino acids
- For all types of sequence data, it can be said that the?more similar?the sequences, the?more closely related?the species are
- Two groups of organisms with very similar sequences have?separated into separate species more recently?than two groups with less similarity in their sequences
- Species that have been separated for longer have had a greater amount of?time to accumulate mutations and changes?to their DNA, mRNA and amino acid sequences
- Sequence analysis and comparison can be used to create family trees that show the?evolutionary relationships?between species
Exam Tip
You may be wondering why you would use amino acids when you could look at DNA or mRNA; it is often easier to find and isolate proteins from cells than it is to isolate DNA or mRNA.However, DNA or mRNA analysis is often more powerful because genes for the same protein may have slightly different base sequences in different species.
Divergence from a Common Ancestor
- The?evolutionary relationships?between species can be determined by analysing?sequence data?from e.g.?DNA, mRNA, or?amino acids in polypeptides
- The?number of differences?between sets of sequence data provides information on?how closely related?two species are
- The?more differences?there are between the sequences, the?longer ago the species diverged, and vice versa
- The differences between sequence data can also be used to produce a?quantitative estimate?for?how long ago?two species diverged from each other
- Differences in sequence data come about due to?mutations?in the DNA
- Evidence suggests that?mutations occur at a constant rate
- This means that the?number of mutations?that have occurred gives an indication of the?amount of time that has passed?since two species diverged
- Scientists refer to the constant rate of mutation as the?molecular clock
- Analysing the differences in sequence data allows evolutionary biologists to determine the?order?in which different species?diverged from a common ancestor, and therefore?how closely related?species are

Differences in DNA sequence data show how much time has passed since species diverged from each other, enabling the relationships between species to be established
Analogous & Homologous Traits
- Homologous?traits?can be defined as
- Characteristics that may differ in form and function in different species but that have shared evolutionary origins
- Homologous traits, or characteristics,?indicate common ancestry, and are useful for classifying organisms into?true clades
- An example of a homologous characteristic is the?pentadactyl limb; limbs in different species of animal differ significantly in their shape and role, but similarities in overall structure indicate common ancestry
- The difficulty with using homologous traits in classification is that it is not always obvious whether characteristics are?homologous?or?analogous
- Analogous traits?can be defined as
- Characteristics with the same function but which do not share an evolutionary origin
- Such characteristics have?evolved independently of each other?from?different ancestors, enabling organisms to?adapt to similar environments
- This is known as?convergent evolution
- Analogous characteristics?look similar, hence the danger of confusing them with homologous characteristics
- Classifying organisms on the basis of analogous characteristics will?not produce an accurate clade
- This has led to?errors?of classification in the past
- For this reason,?sequencing data?is now used for classification instead of observable characteristics
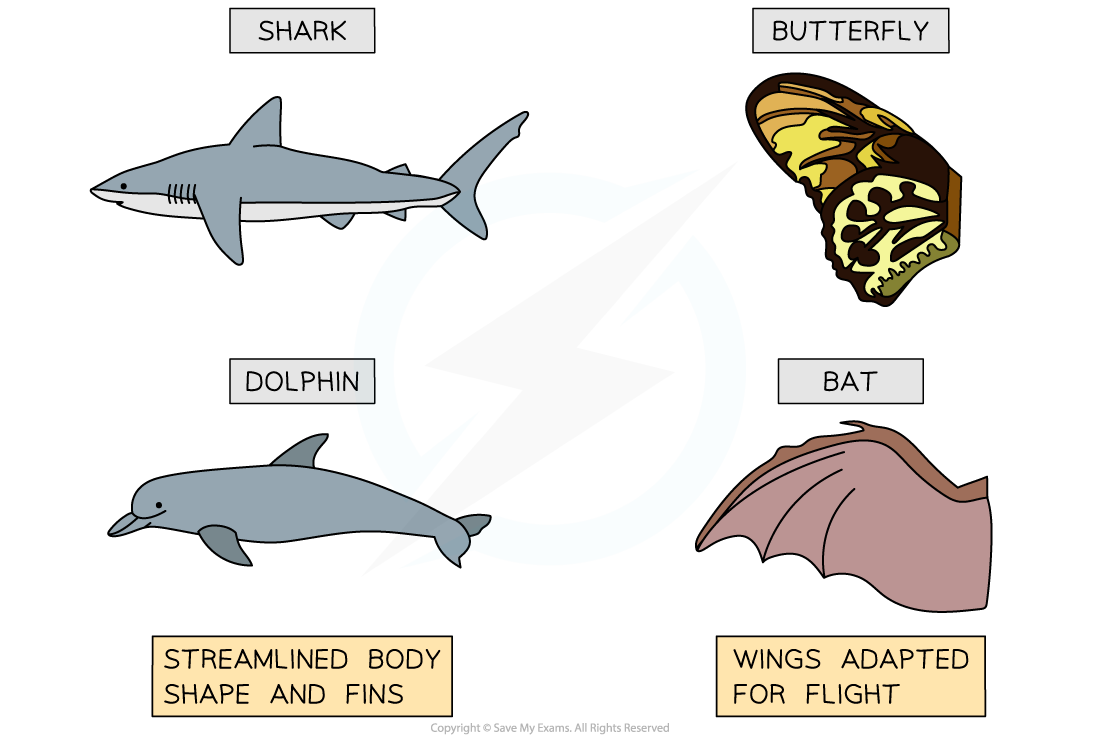
The body shape of sharks/dolphins and the wings of butterflies/bats are both examples of analogous structures
Cladograms
- Evolutionary relationships?between species can be represented visually using a diagram called a?cladogram
- Cladograms are?evolutionary trees?that show probable?order of divergence from ancestral species?and therefore probable?relationships between species
- The point at which two branches separate is known as a?node
- Nodes represent?common ancestor?species
- The information used to build cladograms most often comes from?sequence data?due to difficulties in the use of observable characteristics
- It can be difficult to be sure whether observable traits are?homologous?or?analogous
- Sequence data can provide information about?how different species are?from each other, as well as?how much time has passed?since divergence from a common ancestor took place
- The constant rate at which mutations accumulate can be used as a?molecular clock
- Computers?use the information from sequence data to?build the most likely cladogram
- This is done using the principle of?parsimony, which states that the simplest explanation is preferred
- The computer builds the?shortest possible cladogram?with the?smallest number of divergence events?to fit the available data
- We say that cladograms show the?most probable?divergence times and relationships rather than providing?definite?conclusions
- This is done using the principle of?parsimony, which states that the simplest explanation is preferred
Cladograms that include humans and other primates
- Analysis of?sequence data?for humans and other primate groups show that?humans are most closely related to chimps and bonobos, and that the?next closest relative is the gorilla
- Humans are thought to have?diverged?from chimps and bonobos between 5-7 million years ago
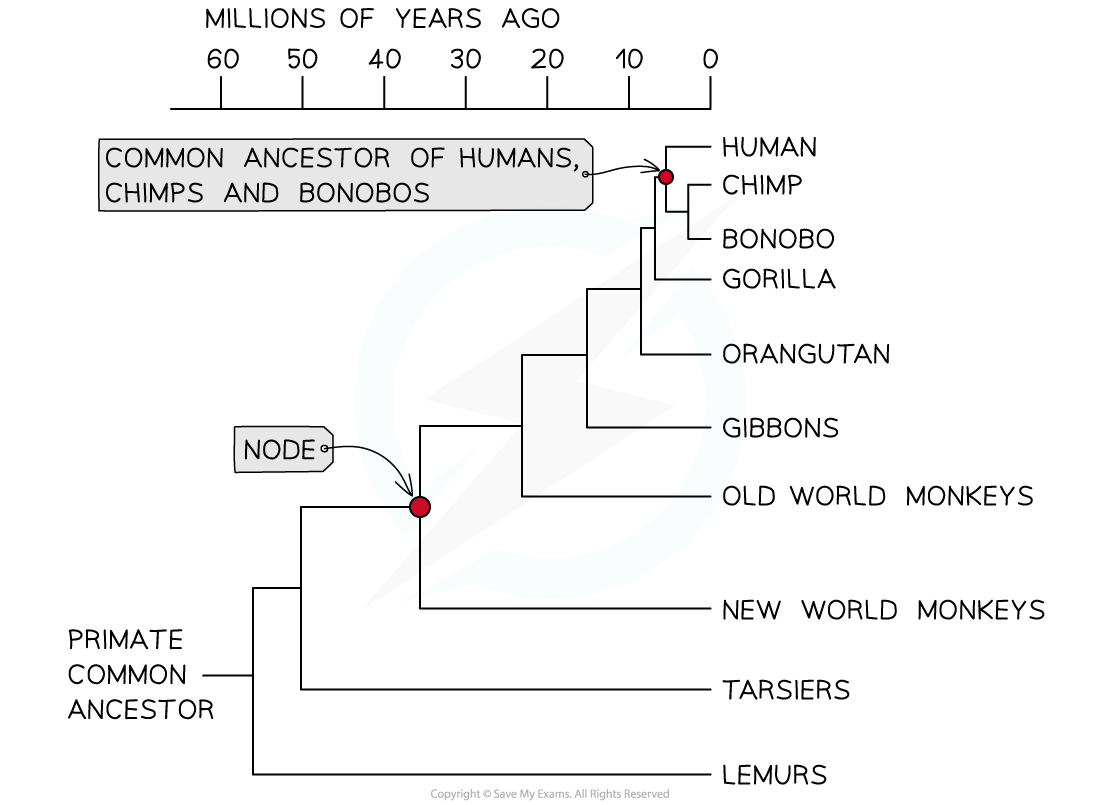
A cladogram showing humans and other primates
轉載自savemyexams

最新發布
? 2025. All Rights Reserved. 滬ICP備2023009024號-1









